Glencoe Software and the OME team are pleased to announce a new collection of command line tools for converting whole slide images to pyramidal OME-TIFF.
Glencoe Software and the OME team are pleased to announce a new collection of command line tools for converting whole slide images from proprietary file formats into pyramidal OME-TIFF; an open file format. These tools were initially designed to convert Philips’ iSyntax and 3DHISTECH’s .mrxs file formats, but can also be used with any other whole slide format supported by Bio-Formats. Our aim is to use these tools to help further the aims of interoperability and openness in digital pathology data.
Why convert?
For most formats, we have been able to write an open source Bio-Formats reader that is reasonably fast and does not rely upon a third-party SDK. In the case of iSyntax, using Philips’ SDK was necessary as reverse engineering the format was not practical. This SDK is publicly distributed under a restrictive license that prevents redistribution, making it difficult to include in applications such as OMERO, QuPath and Fiji. The SDK is also constructed using Python with a C++ core, consequently there are currently no Java bindings. Significant effort would have been required to use the SDK within Bio-Formats. In the case of .mrxs, we were able to create a Bio-Formats reader that could read the largest image in the pyramid, but not the downsampled pyramid levels. The tile sizes, layout, and compression all mean that this reader is quite slow to read image data.
These are examples of several issues we have seen come up across the proprietary file format ecosystem over the last couple of years. Across several imaging domains, our experience is that many proprietary file formats are poorly suited for the multiple writer / multiple reader parallel programming model that so often accompanies the cloud computing workflows that drive AI and deep learning in biological imaging. Several projects1,2,3,4, including OME, are pursuing data storage strategies that are compatible with these workflows. Clinicians, experimentalists, data scientists, and IT professionals need to be able to store data in a way that makes sense to them, is performant, portable and cost effective. This is not necessarily compatible with the way the instrument wrote the data to disk in a proprietary file format during acquisition, making conversion a necessity.
The solution
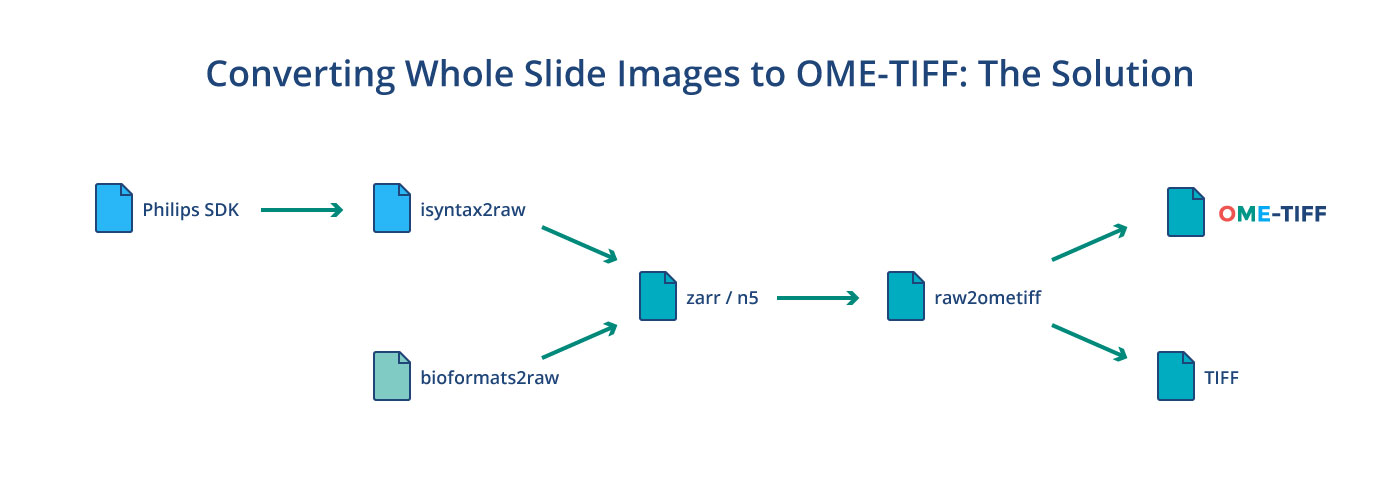
For both the Philips iSyntax and 3DHISTECH .mrxs proprietary file formats we use a two step process to convert to OME-TIFF. The first step is a format-specific application that converts to a temporary N5 or Zarr structure. For iSyntax, this is isyntax2raw, which is a Python 3 application that uses Philips’ SDK directly and for .mrxs, this is bioformats2raw, which is a Java application that includes a separate reader for .mrxs that does not ship with Bio-Formats. In both cases, the slide is broken up into tiles which are processed in parallel, downsampled and finally losslessly compressed to create a temporary image pyramid. Tile sizes, compression as well as file format are configurable putting these choices in the hands of clinicians, experimentalists, data scientists, and IT professionals. By default, N5 with LZ4 compression is used. We chose these over HDF5 or multipage TIFF primarily because of the multiple writer / multiple reader programming model these formats support. If additional information such as slide labels, overview images, or acquisition metadata was in the original file, then this will be converted as well using OME-XML as the intermediary.
The Philips iSyntax SDK is required for any iSyntax file conversion to take place. It must be downloaded separately from Philips and the relevant license agreement agreed to.
Once the initial conversion to a temporary pyramid is finished, the raw2ometiff command line tool is used to convert to a pyramidal OME-TIFF or a plain TIFF that can be read by other software. raw2ometiff uses Bio-Formats’ low-level TiffSaver API to perform compression and direct writes to the same output file on disk using multiple threads. It can do this because all the concurrent heavy lifting has already been done during the first phase. Just like the first step, compression is completely configurable. All the compression types supported by Bio-Formats, such as JPEG-2000, JPEG, and zlib can be used. The converted OME-TIFF file can then be imported in any application that uses Bio-Formats 6.0.0 or later, including OMERO 5.5.0+, QuPath and Fiji/ImageJ.
isyntax2raw is available under the permissive BSD license. bioformats2raw and raw2ometiff are available under the GPL.
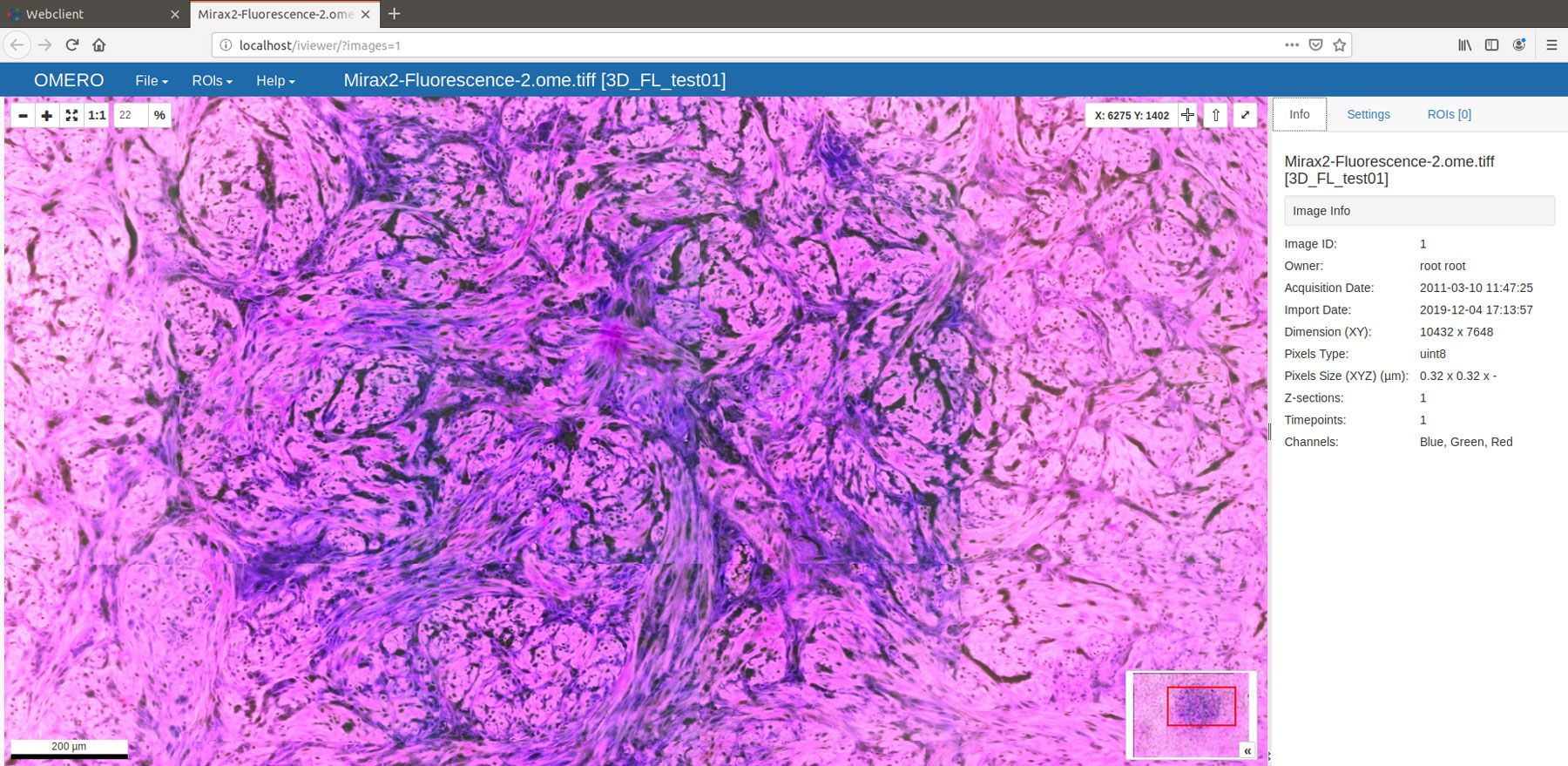
Figure 1: Viewing a freshly converted fluorescence .mrxs file using OMERO 5.5.1 and OMERO.iviewer 0.8.1
Both conversion steps were explicitly designed to take advantage of high-performance computing environments when possible. Supplying more memory and CPUs will result in faster conversion times, which may be as little as 5 minutes in total for 40 or more GiB of pixel data. Exact conversion times vary based upon the hardware and command line options used, as well as the input image’s format and number of pixels. We encourage experimenting with different settings using OME’s public datasets and test data from Openslide.
This two step design will also allow other future conversion targets, such as DICOM, to use N5 or Zarr as the source rather than the original proprietary image file. The current N5 or Zarr intermediate format should be considered temporary at this stage of development as it is likely to undergo several changes over the coming months. As the communities around storing image data in these formats evolve we expect the conversion to OME-TIFF to become unnecessary.
We look forward to your feedback and comments. Our thanks to iCAIRD, PathLAKE, and InnovateUK for the support that made this work possible.
1 TileDB (https://www.tiledb.io/)
2 Zarr (https://zarr.readthedocs.io/en/stable/; https://www.bdi.ox.ac.uk/news/zarr-project-to-receive-funding-from-the-chan-zuckerberg-initiative)
3 N5 (https://github.com/saalfeldlab/n5)
4 KLB (https://bitbucket.org/fernandoamat/keller-lab-block-filetype/src/master/)
