In a recent workshop on tissue clearing, hosted at the Harvard Center for Biological Imaging, we spoke about considerations for the structure of imaging data within the OME-Zarr file format which can impact analysis and visualization workflows, in particular for multi TB lightsheet or other 3D imaging acquisitions.
We have written previously about the advantages of working with Next-Generation File Formats such as OME-Zarr for bioimaging workflows. In particular, OME-Zarr improves interoperability and scalability of analysis and visualization workflows, due to its open specification, open source parallel readers and writers, and support for arbitrary, chunk-wise data access. Here we detail further some considerations for the structure of imaging data within this OME-Zarr file format which can impact analysis and visualization workflows, in particular for multi TB lightsheet or other 3D imaging acquisitions.
Background
OME-Zarr differs from common monolithic file formats in that it instead separates image data into separate files, so-called chunks. Open source converters which write OME-Zarr, such as bioformats2raw, allow the configurability of this chunk size, including the option to generate 2 or 3 dimensional chunks. In order to make such a decision, the downstream workflows for this data must be considered.
Workflow examples - image viewers
OMERO Plus viewers such as PathViewer are planar, meaning they display only the XY view of the data, rather than generating orthogonal or volume views, even for 3D data. Because of this design, the data loaded for any given view of the image will never be beyond a single Z-plane. 3D chunking is therefore suboptimal when compared to 2D chunking.
WEBKNOSSOS is an open-source tool for exploring large 3D datasets as well as collaboratively generating annotations, and it was chosen for this example due to its shared compatibility with the OME-Zarr format. In contrast to PathViewer, WEBKNOSSOS generates orthogonal views of 3D data on the fly, meaning any given XY view of the data will be accompanied by XZ and YZ views, which must be calculated from across the X, Y and Z dimensions. 3D isotropic chunks are optimal in this case because all three dimensions are represented in each chunk and can therefore be used in orthogonal view computations.
Results
The graph below illustrates a core concept of chunk shape impacting the read speeds for bioimaging data. Two copies of the same dataset were generated with different chunk dimensions using bioformats2raw, and both were stored in and accessed via Amazon’s S3. The chunk dimensions shown, labeled as “2D chunking” and “3D chunking” are specifically 1024 x 1024 x 1 and 32 x 32 x 32, respectively. These dimensions were chosen because they are the default or preferred chunk dimensions for OMERO Plus and WEBKNOSSOS, respectively. Various array subsets were read using the zarr-py library and the timings of these reads were compared between the two datasets. What is clear from these numbers is that loading 2D data is faster with 2D chunking, but loading 3D data or orthogonal data is faster with 3D chunking.
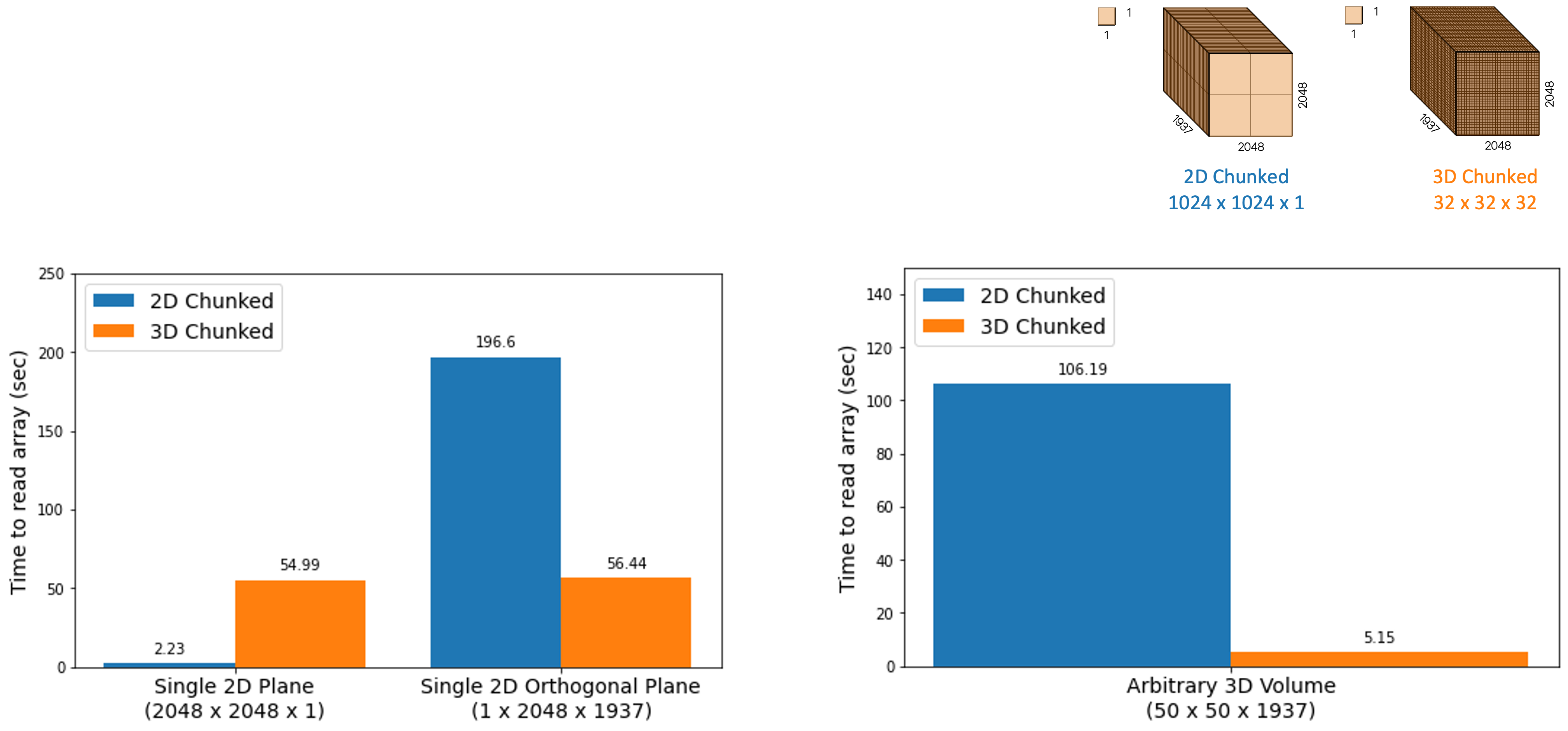
Note that the measurements above did not consider parallel reads or other optimizations, and are only reported as the time to read all data for the selected subset, rather than the time to first byte.
See our public example of reading OME-Zarr data here if you would like to experiment on your own data.
Conclusions
The conclusion from this simple test is clear: there is not a one-size-fits-all solution for data structure, but this structure does dramatically impact downstream visualization and analysis workflows which require performant reading of imaging data. When considering the scalability of visualization and analysis of large datasets, consideration should be applied as early as possible to the formats chosen and the structure of data within. OME-Zarr and other Next-Generation File Formats provide a clear advantage because this choice is given to the user rather than prescribed for a single expected workflow.
Questions or comments? We would love to hear from you. Reach out to us at info@glencoesoftware.com.
