Glencoe Software are happy to announce the latest release of NGFF-Converter. Thanks to our customers and those in the bioimaging community for your use of and feedback on this tool for the conversion of bioimaging data into the open standard formats OME-NGFF and OME-TIFF.
Glencoe Software are happy to announce the latest release of NGFF-Converter.
Proprietary bioimage file formats continue to add unnecessary complexity to image analysis workflows. Conversion to open-source formats such as OME-NGFF and OME-TIFF can make it easier to handle this data. Following the success of the original NGFF-Converter, we’ve listened to user feedback and made some significant changes to improve this utility.
NGFF-Converter 2.0 has been rebuilt from the ground up to provide a more comprehensive interface around the bioformats2raw and raw2ometiff conversion standards. Key changes are as follows:
- User interface has been redesigned and modernised.
- Conversion “Jobs” are now broken down and displayed to the user as a sequence of configurable “Tasks”.
- Settings panels are available for each underlying task, instead of requiring the user to supply command line arguments.
- Progress bars are now shown to the user during conversion.
- Conversion jobs can now be started/stopped individually.
- It is now possible to import/export conversion settings and set user defaults.
- Job settings can now be configured in bulk.
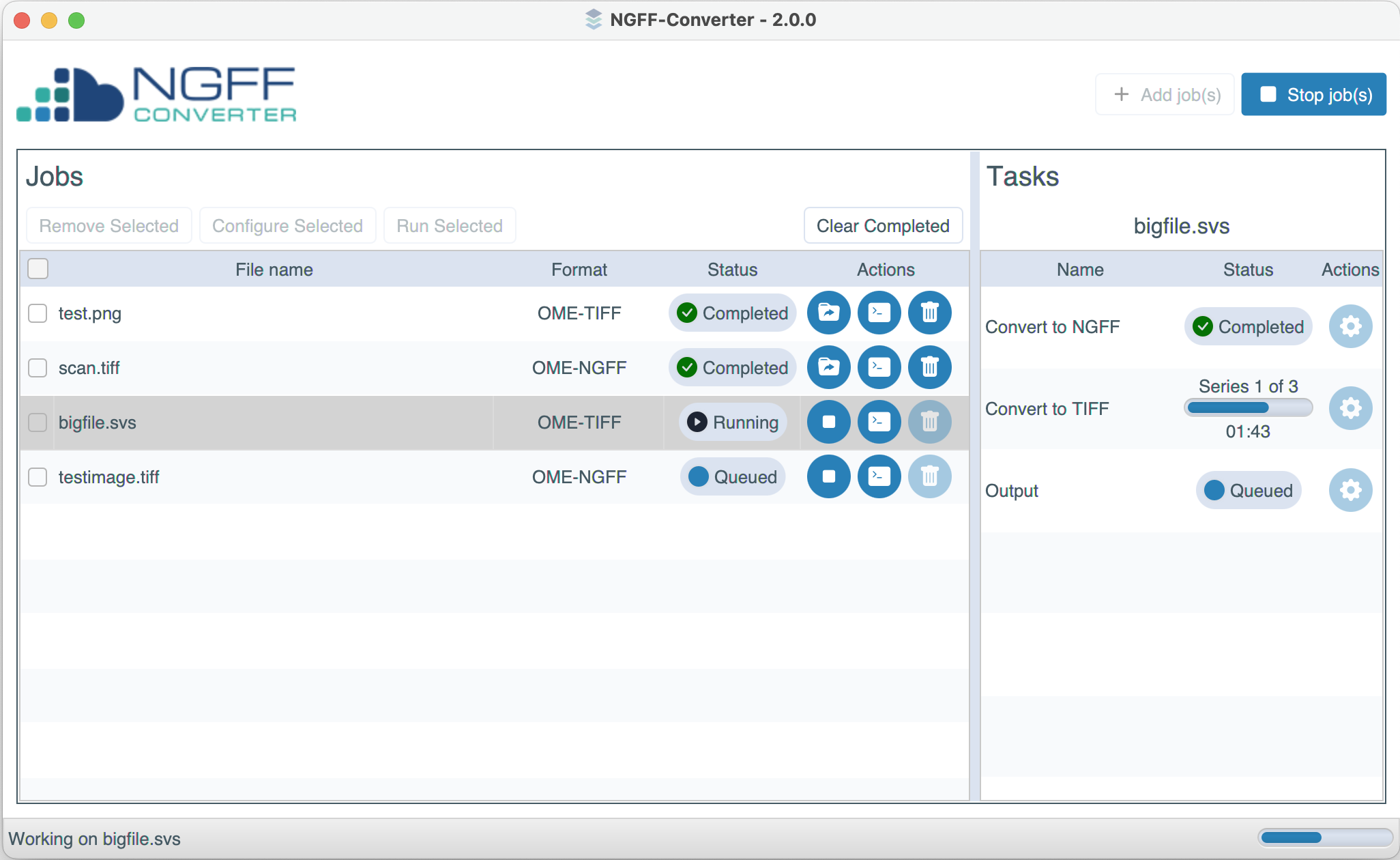 Redesigned user interface for NGFF-Converter 2.0.
Redesigned user interface for NGFF-Converter 2.0.
On the back-end, improvements to the underlying conversion packages (bioformats2raw and raw2ometiff) have allowed for closer integration than was possible with the 1.x series. The utility is also now modular in design, which will simplify adding new conversion workflows and features in future versions.
A huge thanks goes out to all the users who sent us their feedback on the original tool - we hope that this release will deliver a smoother experience that satisfies both new and experienced users.
Read more about NGFF-Converter here.
