At this year’s SBI2 conference in Boston, Glencoe Software are happy to present our work towards a new suite of tools for OMERO Plus aimed at managing complex analysis workflows within OMERO Plus.
Expanding OMERO Plus for orchestration of complex analysis workflows
OMERO Plus provides a comprehensive platform for storage, management and access to image datasets, coupled with the professional support and warranty required for enterprise-level deployments. With existing technologies for scaling and data visualization and new features for analysis and automation presented here, OMERO Plus becomes an orchestration platform of choice for complex and flexible analysis routines at the world’s largest organizations.
High throughput imaging allows scientists to develop increasingly sophisticated image-based assays composed of large volumes of both image data and metadata. As analysis workflows have become more sophisticated, users have been increasingly interested in automating these processes within the OMERO Plus ecosystem. At this year’s SBI2 conference in Boston, Glencoe Software are happy to present our work towards a new suite of tools for OMERO Plus aimed at addressing this growing challenge.
Management of Experimental Results
While OMERO Plus offers first-class metadata management, we have noted that users are increasingly producing large quantities of file attachments as the output of analysis and reporting routines (CSVs, PDFs, etc.), which can be difficult to review and explore using the existing OMERO.web interface. Aiming to address this, we have developed a new plugin to introduce a dedicated “Analysis” tab into the web client. This will provide a comprehensive interface organizing these results, grouped by each analysis run. In addition to providing intuitive access to data from a specific analysis, the panel offers convenient links to download results and launch other visualization tools. This panel also provides controls for initiating new analysis workflows from within the OMERO.web environment.
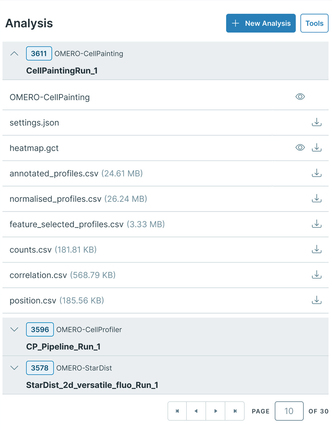
Monitoring of Multi-Stage Workflows
The OMERO.scripts interface can be leveraged to launch third party tooling or submit large computational jobs to high performance computing (HPC) environments. While this provides a convenient means of initiating an analysis from within OMERO Plus, the resulting HPC or cloud jobs previously had no representation within OMERO Plus interfaces. It was therefore impossible for users to troubleshoot or check the progress of their workflows from within the OMERO Plus interface.
To address this, we have implemented a system for representing these processes within the OMERO Plus data model as “Workflows”. Each workflow consists of multiple “Tasks” to represent individual steps in the process. External tools are able to report progress and results back to OMERO Plus through a simple API, keeping the user up to date with the workflow status. Analysis results are also associated with these Workflow objects and provide traceability between the input settings and resulting data.
To provide the users with an interface for checking the status of Workflows, an additional new OMERO.web plugin will offer an overview of both active and past submissions. Each workflow’s individual steps can be reviewed and monitored in detail. Complex tasks can also be represented with progress bars, providing the user with a visual representation of task status. In addition to offering information on status, the interface also displays links to useful outputs such as logs and error messages to aid troubleshooting.
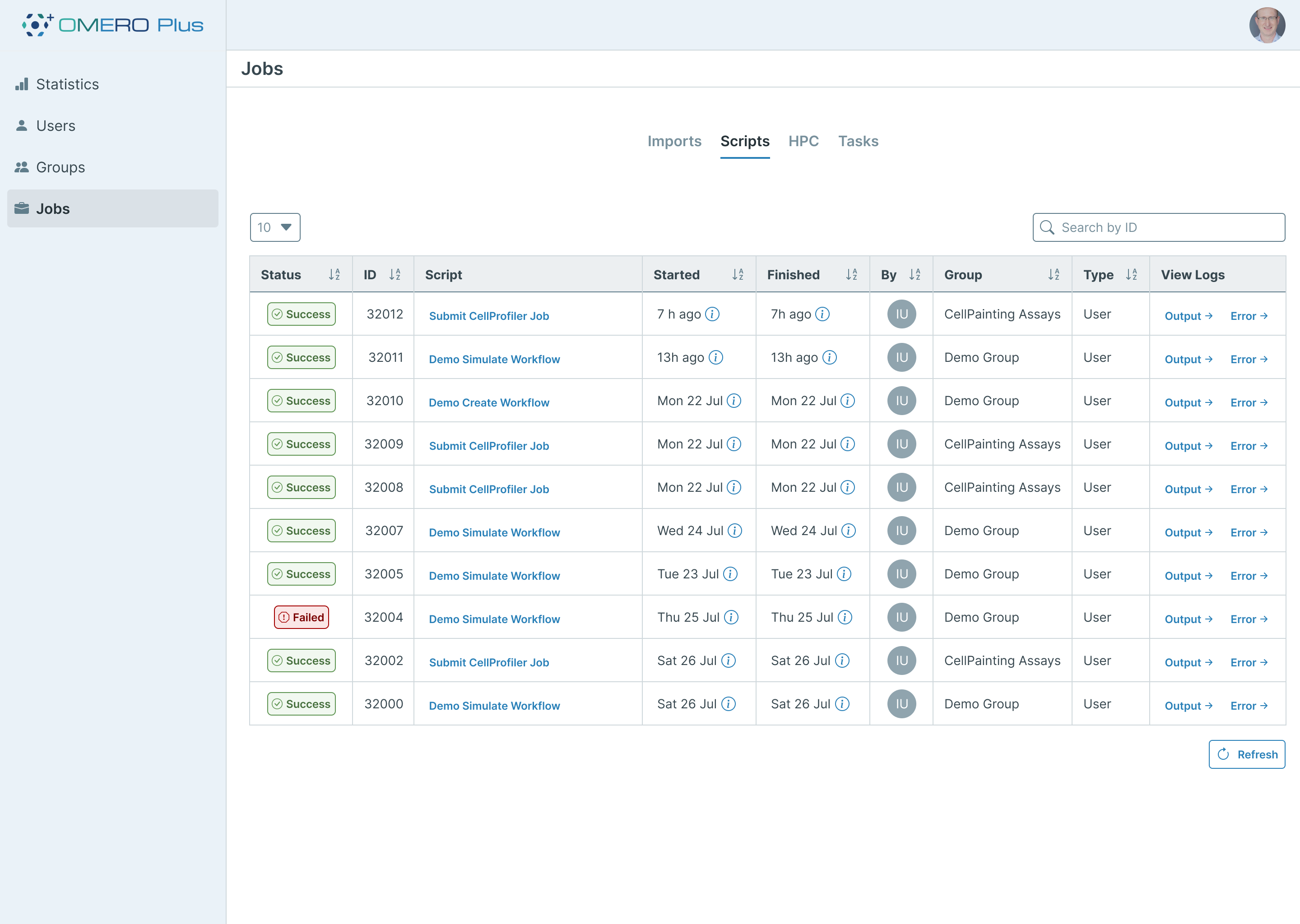
In addition to complex analysis routines, this interface can also be used to monitor standard OMERO.scripts and import jobs, providing a unified, searchable interface for any long running operations which execute on an OMERO Plus system.
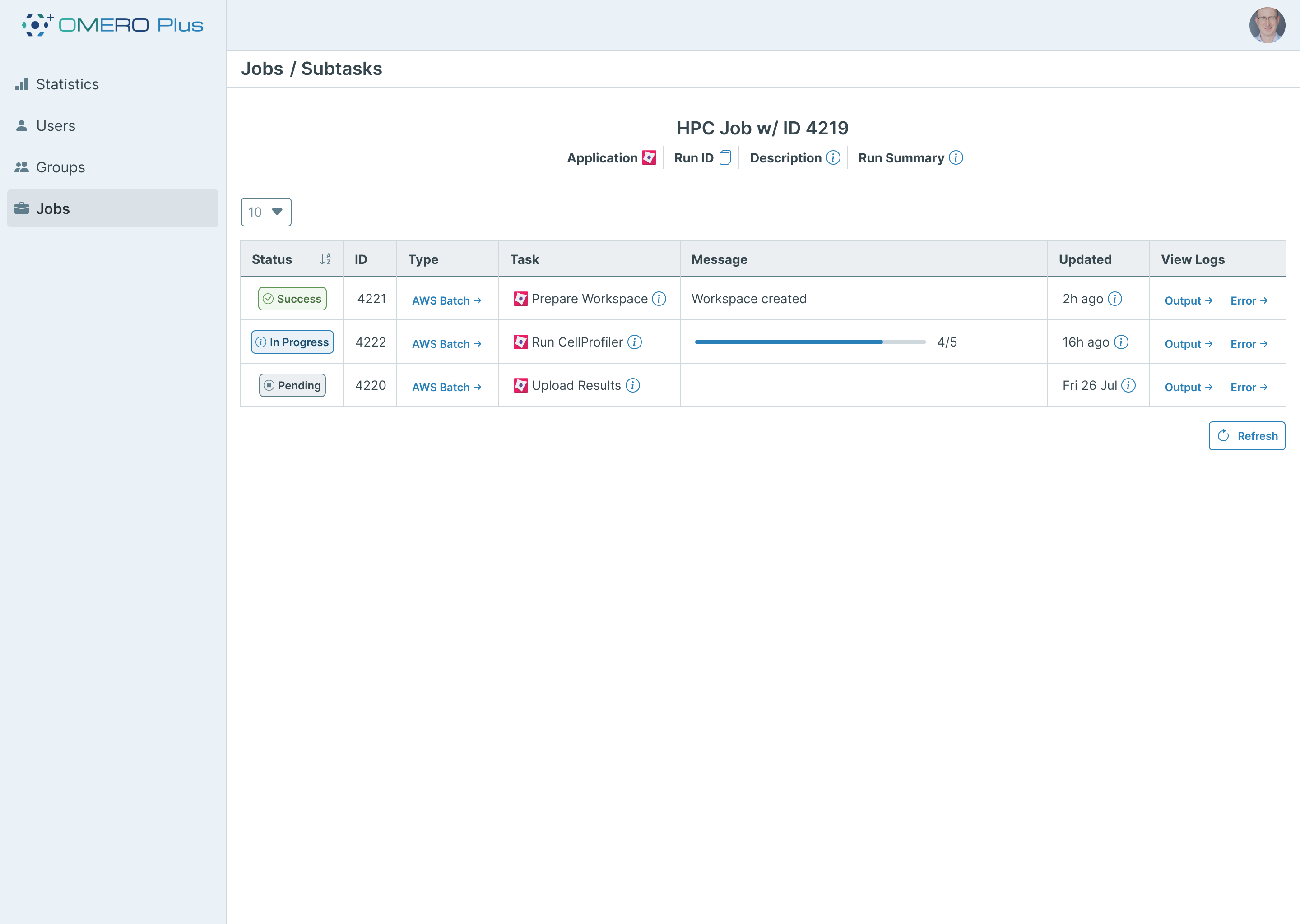
Guided Configuration of Analysis Workflows
While OMERO.scripts offer limited configurability of inputs, many modern analysis protocols permit extensive customization and parameter tuning. A standard practice is the use of complex configuration files, requiring an expert user to create or edit, and which contain too many options to be interpretable within the existing OMERO.scripts interface. To address this, we aim to introduce new tools that offer a more complete and user-friendly experience for guiding the execution of complex technical workflows.
As a case study, we have investigated methods for performing analysis on Cell Painting data. Cell Painting is a high content screening assay which involves staining cells for multiple compartments and collecting an extremely diverse array of measurements for individual cells. Downstream analysis then produces “profiles” of typical cells under each experimental condition and isolates the specific measurement features which are impacted by the treatments (feature reduction).
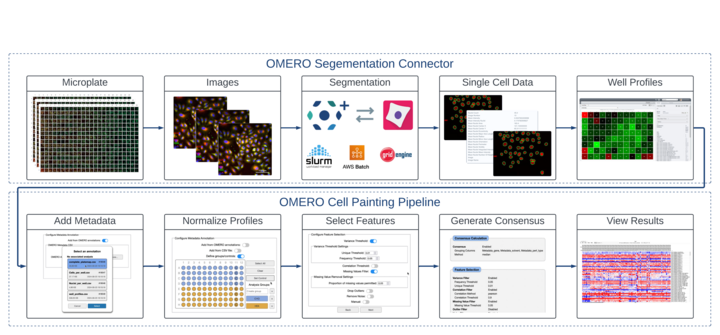
Schematic of the OMERO Plus-based Cell Painting analysis workflow
Glencoe Software’s OMERO Segmentation Connector provides the ability to launch CellProfiler analysis from within OMERO Plus. Whilst this Connector is capable of performing the intensive single-cell detection and quantitation for the first part of the Cell Painting workflow, performing feature reduction and analysis previously remained a manual downstream process performed outside of OMERO Plus.
The analysis steps to convert the well-level profiles produced by the OMERO Segmentation Connector into human-interpretable results involve extensive configuration options, and so have typically been performed in custom Jupyter notebooks. The Jupyter environment operates as a series of blocks of Python code, restricting users without a computational background from being able to perform downstream analysis of their data. Furthermore, analysis settings are usually manually configured inline by the user. This poses a challenge for ensuring reproducibility and traceability of the resulting output data.
To address these challenges, we have developed an OMERO Plus extension which guides users through configuration and submission of Cell Painting analysis workflows. This tooling provides an interactive, modular interface available within the OMERO.web client. Allowing configuration in such an extension allows us to provide dedicated widgets for choosing settings which can otherwise be difficult to configure. For example, defining experimental controls and groups can now be performed within an intuitive visual plate widget.
The extension also breaks down the large list of configuration options into a series of pages, which the user is guided through with appropriate help tooltips. In this interactive interface the user is only presented with configuration options which are relevant to the workflow as currently configured. For instance, the Cell Painting workflow offers multiple feature reduction options such as filtering for correlation, variance or even manually including or excluding measurements. The reactive interface ensures that an analyst is only exposed to options which are relevant to their experiment.
Once configured, the user reviews their chosen settings before submitting to whichever compute environment is available for their OMERO Plus installation (e.g. SLURM, AWS Batch). This creates a new Workflow which is represented within the previously described interfaces for job monitoring and results exploration. Together with OMERO Plus’s existing image and metadata handling, these tools provide an end-to-end interface for performing analysis and reviewing results.
In the case of Cell Painting tabular results representing typical cell profiles within each experimental condition are produced as outputs. The OMERO.web “Analysis” panel plugin provides helpful links to review this data in the Morpheus heatmap tool, and can be extended with options to launch other tooling as desired by the experimenter.
We also acknowledge that data post processing and visualization pipelines are often custom and evolving. Therefore Glencoe Software also created and maintains the omero2pandas Python package, which allows resulting tabular data, managed within OMERO Plus, to be directly loaded into Python environments for further handling as desired. Experimental data and results are stored within OMERO Plus, but will always remain accessible to other tooling if preferred by the user.
Summary
Together, these new tools will provide a convenient, all-in-one solution for initiating, monitoring and reviewing results from complex analysis workflows. Importantly, the same principles applied to the Cell Painting use case can be applied to other multi-step analysis pipelines as desired by scientists. The ability to offer a reactive interface for choosing analysis settings provides an improved user experience which also opens up these techniques to scientists without a computational background. If you have a computational workflow which would benefit from such guided configuration, we encourage you to reach out with the contact form below.
Acknowledgements
For demonstration we used the dataset cpg0000 (Chandrasekaran et al., 2022), available from the Cell Painting Gallery on the Registry of Open Data on AWS.
