Glencoe Software is pleased to announce the latest release of OMERO Plus, highlighting open source contributions for the handling of large analytical datasets within OMERO.tables and enhanced support for image archival, as well as new high-content format support.
To see specific version information of all components, see the section below.
OMERO.tables
OMERO Plus has a suite of tools to manage analytical data alongside image data, not only for greater traceability but also to enable easily shareable browser-based visualizations and data mining. In particular for Glencoe Software’s PathViewer and Pageant, efficient interaction with large tabular datasets, stored as OMERO.tables, is essential. Routinely, these tables contain millions of rows (representing cells, nuclei, etc.) and hundreds of columns (measurements, classifications, features, expression levels, etc.). This latest release of OMERO Plus includes substantial performance improvements for the management and mining of large tabular analytical data.
There are two major categories of interactions with OMERO.tables in PathViewer and Pageant:
- Query the table to return matching object identities. In practice, this could mean returning the cells which belong to a certain class in order to filter them in or out of a visualization.
- Return an entire column of data. In practice, this could be used to generate a heat map, plot, or other raw data visualization.
For each of the above scenarios, Glencoe Software has made two major API contributions to open source OMERO.web:
- For querying:
/webgateway/table/<table ID>/rows/accepts a query argument in the usual format and returns matching table row numbers. No actual data from the table is returned. The previous strategy used the/webgateway/table/<table ID>/obj_id_bitmask/endpoint to retrieve a 0 or 1 for all rows in the table. - For raw data retrieval:
/webgateway/table/<table ID>/slice/takes numeric lists of rows and columns and performs a slice. Data is returned in columnar format (rather than rows as in the current table API).
See performance benchmarking in the plots below, matching the two scenarios described above for 0.5 million data points (out of a total table size of 1.2M rows and 100 columns).
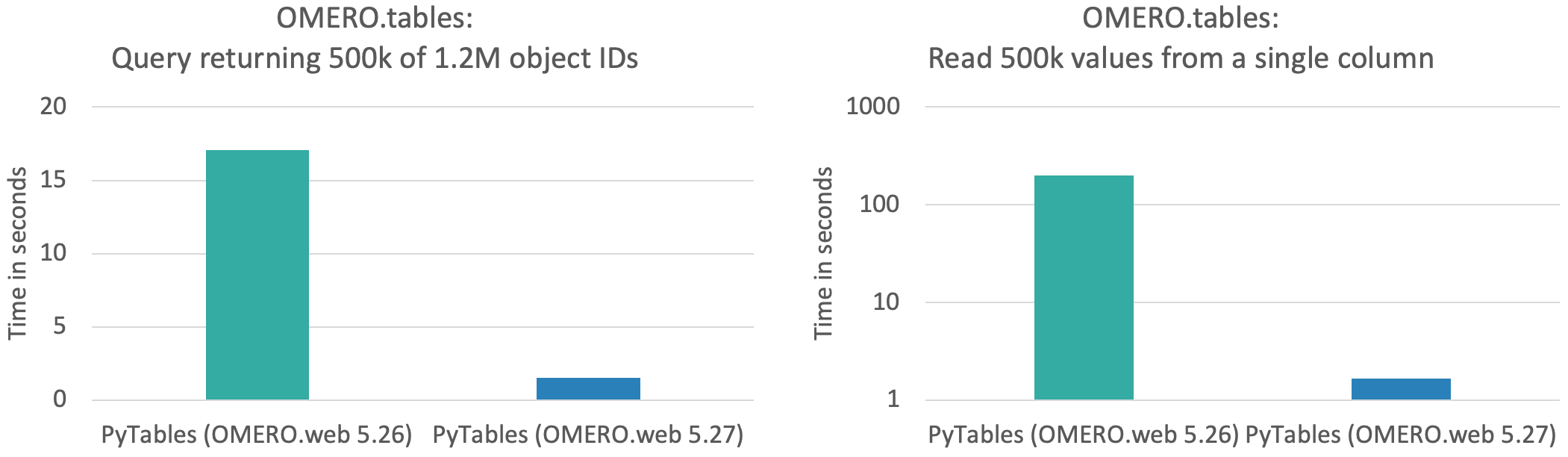
Read more and see the contributions here.
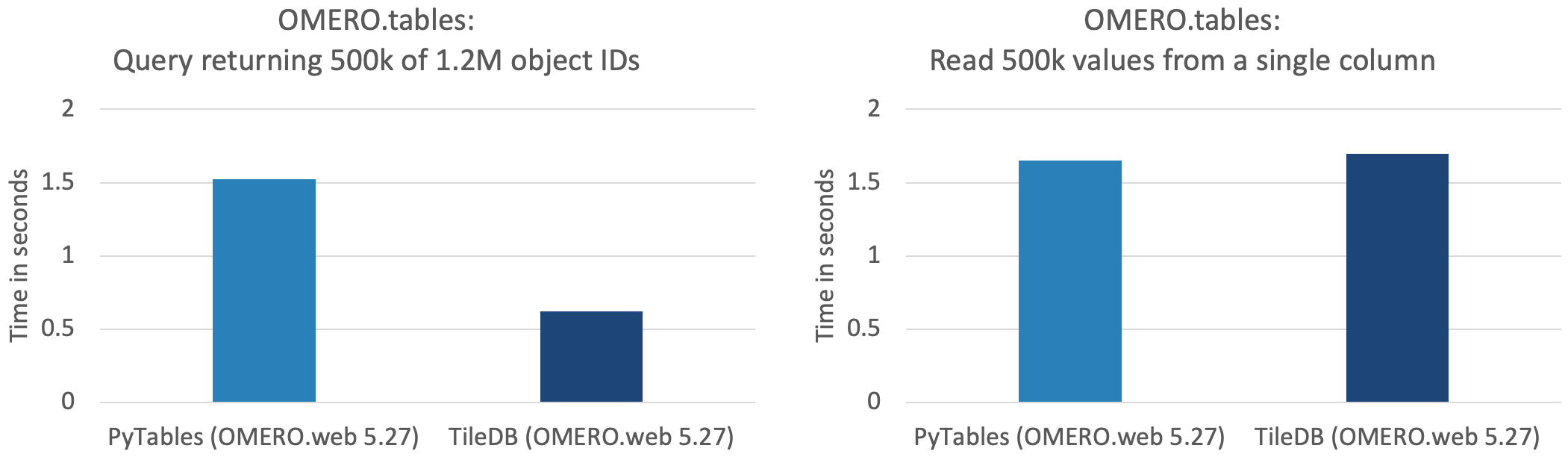
In addition, Glencoe Software has implemented a new backend for OMERO.tables in OMERO Plus, which uses TileDB, rather than PyTables, as the tabular data store. Again, see performance benchmarking in the plots below, matching the two scenarios described above for 0.5 million data points (out of a total table size of 1.2M rows and 100 columns).
Archival support
Although the OMERO data model has long supported the setting of an archived status on an image, the latest version of OMERO.web now includes a visual indicator to reflect this status. See the OMERO.web release notes here.
Further, Glencoe has proposed publicly next steps to improve archival support within OMERO and OMERO Plus.
Contact Glencoe Software support to learn how we can integrate with your organization’s preferred method for archival.
Omero2pandas
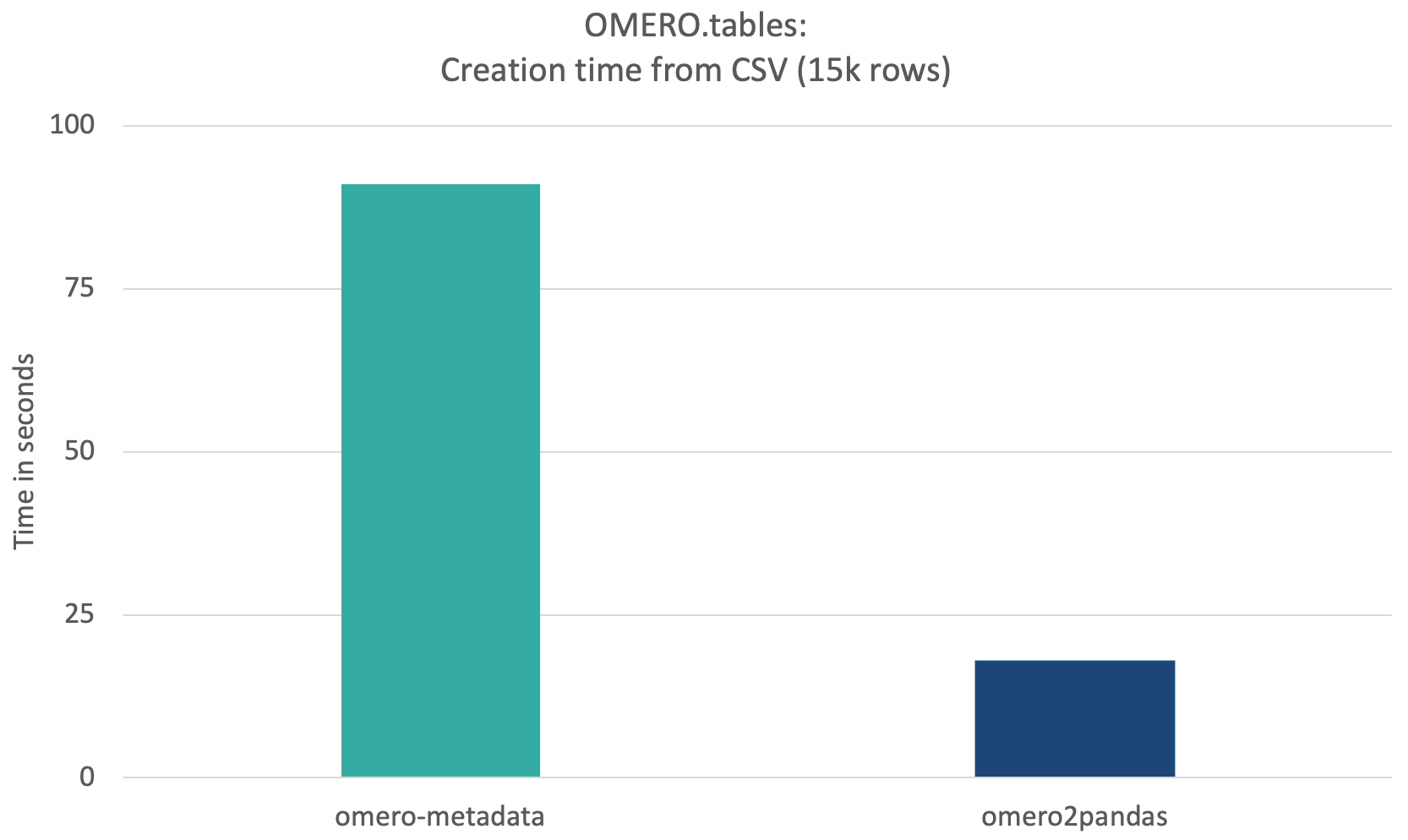
Omero2pandas 0.3.0 has been released! In particular and related to the performance improvements above, omero2pandas now supports the uploading of tables from any CSV file on disk, rather than requiring the user to have a dataframe in memory. Because this upload is also chunked, CSV input files larger than system memory can now be used to create an OMERO.table. Read more about configuring this option here. When compared to other options like omero-metadata, omero2pandas provides a clear advantage in handling large tabular analytical data.
Expert users of this tooling may also note that OMERO.tables can now be linked to multiple objects when created using omero2pandas, including label image ROIs. Linking an OMERO.table to an OMERO Plus label image enables downstream classification workflows in PathViewer and Pageant. Read more here.
With these scalability and usability improvements, omero2pandas has become the standard across Glencoe Software’s tooling for interacting with tabular data, and we hope the same can be said for our customers’ data analysis workflows.
New readers added to Bio-Formats
Thanks to support for the latest Bio-Formats, Incucyte data archives can now be imported into OMERO Plus. In addition, Bio-Formats now supports a direct database integration with Revvity’s Opera Phenix Database (v5), enabling scalable and automated imports from these systems into OMERO Plus. Read more in the Bio-Formats release notes and see other direct database integrations supported by OMERO Plus here.
Versions
- OMERO.server 5.6.13-207
- Glencoe’s Bio-Formats 7.3.1-710
- omero-ms-image-region 0.10.0
- omero-ms-pixel-buffer 0.7.0
- omero-ms-thumbnail 0.5.9
- omero-plus 0.9.0
- OMERO.web 5.27.2
- PathViewer 3.11.0
Upgrading OMERO Plus
To schedule your upgrade of OMERO Plus and take advantage of these new features, reach out to support@glencoesoftware.com.
